Define of Untargeted Lipidomics
Untargeted lipidomics represents a holistic approach to lipid analysis, wherein the entire lipidome of a biological sample is profiled in an unbiased manner. Unlike targeted lipidomics, which focuses on predefined sets of lipid species, untargeted lipidomics aims to capture the entirety of lipid diversity within a sample, encompassing both major lipid classes and rare, elusive species. By employing advanced mass spectrometry techniques coupled with robust data analysis pipelines, untargeted lipidomics offers researchers an unprecedented opportunity to unravel the complexity of lipid metabolism and its implications in health and disease.
Why Untargeted Lipidomics?
Untargeted lipidomics is a transformative approach that addresses key limitations inherent in targeted lipidomics, making it indispensable in modern biological research. Several compelling reasons underscore the importance and relevance of untargeted lipidomics:
Exploration of Lipid Diversity
One of the primary motivations behind untargeted lipidomics is the exploration of lipid diversity within biological systems. Unlike targeted approaches, which are confined to predefined sets of lipid species, untargeted lipidomics casts a wide net, capturing a broad spectrum of lipid classes, subclasses, and molecular species. This comprehensive coverage extends beyond canonical lipid pathways, allowing for the detection of novel lipid species, isomeric forms, and structurally diverse molecules that may play crucial roles in cellular physiology and pathology.
Discovery Potential
The unbiased nature of untargeted lipidomics unleashes its true discovery potential, enabling researchers to uncover previously unrecognized lipid species and metabolic pathways. By surveying the entire lipidome, untargeted lipidomics has the capacity to reveal unexpected metabolic intermediates, lipid-derived signaling molecules, and regulatory pathways that would have otherwise remained concealed. This discovery-driven approach fosters innovation and opens new avenues for understanding the complex interplay between lipids, metabolism, and cellular function.
Identification of Biomarkers and Signatures
Untargeted lipidomics holds promise for biomarker discovery and the identification of lipidomic signatures associated with physiological states, disease progression, and therapeutic interventions. Through comparative analysis of lipid profiles between healthy and diseased conditions, untargeted lipidomics can unveil distinct lipidomic patterns indicative of disease onset, progression, or response to treatment. These lipidomic signatures serve as valuable diagnostic markers, prognostic indicators, and therapeutic targets in various diseases, including metabolic disorders, cardiovascular diseases, neurodegenerative disorders, and cancer.
Systems-Level Insights
By integrating untargeted lipidomic data with other omics datasets (e.g., genomics, transcriptomics, metabolomics), researchers can gain comprehensive insights into the systems-level regulation of lipid metabolism and its implications for cellular function and disease pathogenesis. This multidimensional approach enables the elucidation of complex molecular networks, metabolic pathways, and regulatory mechanisms governing lipid homeostasis, lipid signaling, and lipid-lipid interactions. Such integrative analyses provide a holistic understanding of biological systems and facilitate the identification of novel therapeutic targets and personalized treatment strategies.
Flexibility and Adaptability
Untargeted lipidomics offers flexibility and adaptability to accommodate diverse experimental designs, sample types, and research objectives. Unlike targeted approaches, which require a priori knowledge of lipid targets and specific assay development, untargeted lipidomics can be applied to exploratory studies, hypothesis generation, and biomarker screening in a wide range of biological contexts. Moreover, advancements in mass spectrometry instrumentation, data acquisition strategies, and computational tools continue to enhance the sensitivity, resolution, and throughput of untargeted lipidomic workflows, making them increasingly accessible and cost-effective for researchers across various disciplines.
Untargeted Lipidomics Workflow:
1. Sample Preparation: This typically involves lipid extraction from biological samples using various solvents like chloroform/methanol or other methods such as liquid-liquid extraction or solid-phase extraction.
2. Mass Spectrometry Analysis: After extraction, the lipid extracts are subjected to mass spectrometry analysis, often coupled with chromatographic separation techniques like liquid chromatography (LC) or gas chromatography (GC). In untargeted lipidomics, the mass spectrometer is set to acquire data across a wide mass range, typically covering the mass-to-charge (m/z) ratios of all detectable lipids within the sample.
3. Data Acquisition: Mass spectrometry generates complex data containing mass-to-charge ratios (m/z) and intensity values for all ions detected in the sample. These data are then processed using bioinformatics tools to identify and quantify lipids.
4. Data Analysis: Data analysis involves preprocessing steps such as peak detection, alignment, normalization, and statistical analysis to identify significant differences in lipid profiles among different samples or conditions. This often involves comparing the lipid profiles of different samples based on their mass spectra.
5. Lipid Identification: Untargeted lipidomics aims to identify as many lipid species as possible within the sample. Lipid identification is typically achieved by comparing the experimental mass spectra with reference databases or through fragmentation analysis to elucidate lipid structures.
6. Biological Interpretation: Once identified, the lipid species are further analyzed to understand their biological significance and potential roles in various cellular processes or disease states. This may involve pathway analysis, lipid class distribution analysis, and correlation with other omics data.
How to Analyze Untargeted Metabolomics Data?
Data Preprocessing:
Data Acquisition: Begin by acquiring raw mass spectrometry data from lipidomic experiments using high-resolution mass spectrometers equipped with appropriate ionization techniques (e.g., electrospray ionization).
Data Conversion: Convert raw mass spectrometry data files (e.g., .raw, .mzML) into a standardized format compatible with downstream data analysis software (e.g., mzXML, mzML).
Peak Detection: Employ peak detection algorithms to identify and extract peaks corresponding to lipid features in the mass spectrometry data. Common algorithms include centroiding, deconvolution, and peak picking algorithms.
Feature Detection and Alignment:
Feature Detection: Identify lipid features (e.g., peaks, m/z values) representing individual lipid species or molecular ions within the mass spectrometry data. This step involves determining the retention time, m/z value, intensity, and other relevant parameters for each detected feature.
Feature Alignment: Align lipid features across multiple samples to account for retention time shifts, m/z deviations, and other variations introduced during data acquisition. Utilize alignment algorithms to match corresponding features and generate a consolidated feature table for further analysis.
Data Normalization:
Intensity Normalization: Normalize lipid intensities across samples to correct for variations in sample preparation, instrument response, and other technical factors. Common normalization methods include total ion intensity normalization, median normalization, and internal standard normalization.
Batch Correction: Correct for batch effects or systematic variations introduced during data acquisition by applying batch correction algorithms. This ensures that lipidomic differences observed between samples are not confounded by technical artifacts.
Statistical Analysis:
Univariate Analysis: Perform univariate statistical tests (e.g., t-test, ANOVA) to identify lipid features that are significantly differentially abundant between experimental groups or conditions. Correct for multiple testing using methods such as false discovery rate (FDR) or Bonferroni correction.
Multivariate Analysis: Apply multivariate statistical techniques (e.g., principal component analysis, partial least squares discriminant analysis) to visualize and explore global patterns of lipidomic variation across samples. These methods help identify sample clusters, detect outliers, and uncover latent structures within the lipidomic dataset.
Annotation and Interpretation:
Lipid Annotation: Annotate lipid features by matching their mass-to-charge ratios (m/z) and retention times with reference databases of lipid standards or computational lipid libraries. Utilize lipid annotation software and databases (e.g., LipidBlast, LipidMaps, LIPID Metabolites and Pathways Strategy) to assign putative lipid identities and annotate lipid classes, subclasses, and structural features.
Pathway Analysis: Conduct pathway analysis to elucidate the biological significance of identified lipidomic changes and infer underlying metabolic pathways or cellular processes. Utilize pathway enrichment analysis tools (e.g., MetaboAnalyst, Mummichog, MetScape) to identify overrepresented lipid pathways, metabolic modules, or functional annotations within the lipidomic dataset.
Integration with Other Omics Data:
Cross-Omics Integration: Integrate untargeted lipidomic data with other omics datasets (e.g., genomics, transcriptomics, metabolomics) to gain a systems-level perspective on lipid metabolism and its interactions with other cellular processes. Utilize bioinformatics tools and integrative analysis pipelines to identify correlations, co-regulated pathways, and network interactions across different omics layers.
Validation and Follow-Up Studies:
Experimental Validation: Validate key findings from untargeted lipidomic analyses through targeted validation experiments, such as quantitative lipid assays, lipidomic profiling using targeted mass spectrometry methods, or functional validation assays in cellular or animal models.
Follow-Up Studies: Design follow-up studies to further investigate novel lipidomic signatures, validate putative biomarkers, or elucidate mechanistic insights into lipid-mediated cellular processes. Utilize complementary experimental approaches, such as lipidomics imaging, lipidomic flux analysis, or lipidomics perturbation studies, to deepen our understanding of lipid biology in health and disease.
Targeted vs. Untargeted Lipidomics
In the realm of lipidomics, two main approaches dominate the landscape: targeted and untargeted lipidomics. While both methodologies aim to unravel the complex lipid composition of biological samples, they differ significantly in their scope, methodology, and applications. Let's delve into the nuances that set them apart:
Targeted Lipidomics:
1. Precision Targeting: Targeted lipidomics involves the precise quantitation and profiling of predefined sets of lipid species or lipid classes. Researchers select specific lipid targets based on prior knowledge, hypotheses, or biological significance, focusing their analytical efforts on these predetermined targets.
2. High Sensitivity and Specificity: Targeted lipidomics assays are optimized for high sensitivity and specificity, allowing for accurate quantitation of targeted lipid species within complex biological matrices. By leveraging targeted methodologies such as selected reaction monitoring (SRM) or multiple reaction monitoring (MRM), researchers achieve robust and reproducible measurements of known lipid analytes.
3. Limited Coverage: While targeted lipidomics offers unparalleled sensitivity and quantitation for predefined lipid targets, its coverage is inherently limited to the selected lipid species or classes. This narrow focus restricts the ability to discover novel lipid species, explore uncharted metabolic pathways, or identify unexpected lipidomic features.
4. Hypothesis-Driven Approach: Targeted lipidomics is hypothesis-driven, relying on prior knowledge or specific research questions to guide lipid target selection and assay development. This focused approach is well-suited for hypothesis-driven research or targeted studies of known lipid pathways or biomarkers.
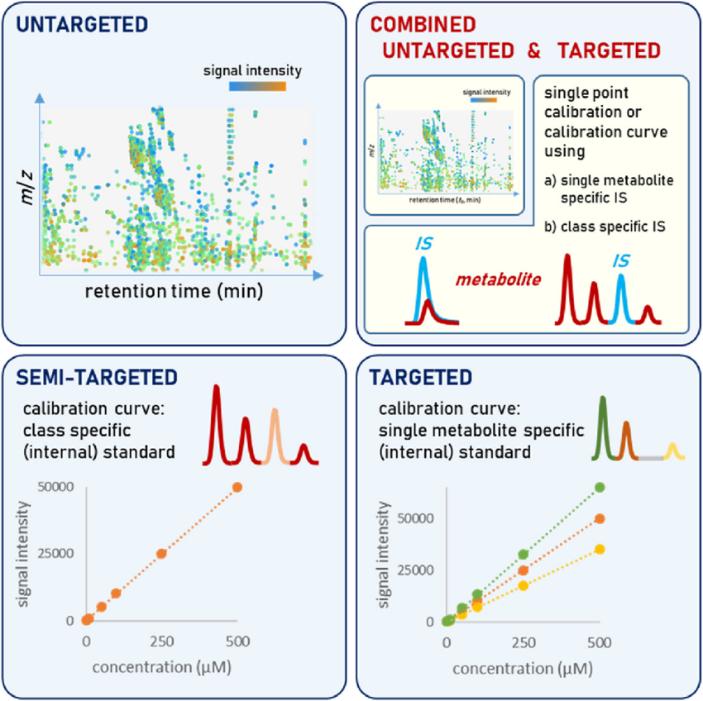
Untargeted and targeted metabolomics and lipidomics methods (Rakusanova et al., 2023).
Untargeted Lipidomics:
1. Comprehensive Profiling: Untargeted lipidomics aims to comprehensively profile the entire lipidome within a biological sample without prior knowledge or bias towards specific lipid targets. This unbiased approach casts a wide net, capturing a diverse array of lipid classes, subclasses, and molecular species present in the sample.
2. Discovery-Driven Exploration: Untargeted lipidomics is discovery-driven, allowing researchers to explore the full spectrum of lipid diversity, uncover novel lipid species, and elucidate unexplored metabolic pathways. By analyzing samples without predefined targets, untargeted lipidomics facilitates the discovery of unexpected lipidomic features and metabolic signatures.
3. Broad Coverage, Sacrificing Sensitivity: While untargeted lipidomics offers comprehensive coverage of the lipidome, its sensitivity and specificity may be lower compared to targeted approaches. The trade-off for broad coverage is the potential for increased noise and decreased sensitivity in detecting low-abundance lipid species or subtle lipidomic changes.
4. Hypothesis-Generating Tool: Untargeted lipidomics serves as a powerful hypothesis-generating tool, providing researchers with a wealth of data to generate hypotheses, formulate research questions, and guide subsequent targeted investigations. By uncovering novel lipidomic signatures and metabolic patterns, untargeted lipidomics drives innovation and opens new avenues for lipidomics research.
Key Differences Summarized:
- Scope: Targeted lipidomics focuses on predefined lipid targets, while untargeted lipidomics explores the entire lipidome in an unbiased manner.
- Approach: Targeted lipidomics is hypothesis-driven, whereas untargeted lipidomics is discovery-driven.
- Coverage vs. Sensitivity: Targeted lipidomics offers high sensitivity and specificity for predefined targets, while untargeted lipidomics sacrifices sensitivity for broad coverage.
- Applications: Targeted lipidomics is suited for hypothesis-driven research or targeted studies of known lipid pathways, biomarkers, or drug targets, while untargeted lipidomics is ideal for exploratory studies, biomarker discovery, and uncovering novel lipidomic signatures.
Applications of Untargeted Lipidomics
Biomarker Discovery:
Untargeted lipidomics facilitates the identification of lipid biomarkers associated with various diseases, such as metabolic disorders, cardiovascular diseases, neurodegenerative diseases, and cancer. Through the profiling of the lipidome in both diseased and healthy cohorts, researchers can discern lipid signatures indicative of pathological states, thereby contributing to early diagnosis, prognosis, and therapeutic monitoring.
Drug Discovery and Development:
In the realm of drug discovery and development, untargeted lipidomics assumes a pivotal role in elucidating lipid-mediated drug mechanisms, evaluating drug efficacy, and discerning off-target effects. By delineating lipidomic alterations consequent to drug interventions, researchers can unveil lipid pathways modulated by pharmacotherapeutics, refine drug candidates, and prognosticate drug responses in both preclinical and clinical domains.
Nutritional Research:
Untargeted lipidomics serves as a potent tool in elucidating lipid metabolism and deciphering the impact of dietary factors on lipid composition, thereby enriching nutritional research. Through the analysis of lipidomic profiles in response to dietary manipulations or nutritional supplementation, researchers can discern lipid biomarkers indicative of nutritional status, evaluate dietary compliance, and unravel lipid-mediated pathways governing metabolic homeostasis.
Environmental Exposure Assessment:
The application of untargeted lipidomics facilitates the evaluation of lipidomic alterations elicited by environmental exposures, encompassing pollutants, toxins, and environmental contaminants. Through the profiling of lipidomic signatures in exposed cohorts or experimental models, researchers can delineate lipid biomarkers reflective of environmental exposure, assess associated health risks, and furnish evidence to guide regulatory initiatives aimed at mitigating environmental hazards.
Personalized Medicine:
Untargeted lipidomics emerges as a promising avenue in the realm of personalized medicine, underpinned by its capacity to delineate individualized lipidomic profiles for the purpose of treatment tailoring. Through the integration of lipidomic data with complementary omics datasets and clinical parameters, researchers can stratify patient cohorts, prognosticate treatment outcomes, and optimize therapeutic interventions to align with the tenets of personalized healthcare delivery.
Functional Lipidomics:
Untargeted lipidomics empowers investigations into the functional roles and interactions of lipids within biological milieus, encompassing lipid signaling, membrane dynamics, and lipid-protein interactions. By delineating lipidomic alterations in response to cellular cues or perturbations, researchers can unravel the functional underpinnings of lipids in cellular physiology, disease pathogenesis, and adaptive responses.
Systems Biology and Network Analysis:
Untargeted lipidomics assumes a pivotal role in systems biology endeavors by furnishing expansive datasets amenable to network analysis, modeling, and simulation of lipid metabolic networks. Through the integration of lipidomic data with other omics datasets and computational methodologies, researchers can reconstruct intricate lipid metabolic pathways, pinpoint pivotal regulatory nodes, and unravel emergent properties inherent to lipid-mediated biological systems.